Most american research labs seek precision when measuring genetic material, yet even small errors can have a big impact in high-stakes fields like cancer diagnostics. With digital PCR, scientists can achieve over one hundred times greater sensitivity than traditional PCR approaches. This technological leap matters because it opens new doors for early disease detection, accurate monitoring, and personalized medicine. Discover how digital PCR sets new standards in genetic analysis and why its core principles are reshaping molecular science.
Table of Contents
- Defining Digital PCR And Its Core Principles
- Comparing Digital PCR, QPCR, And Variants
- Step-By-Step Digital PCR Workflow Explained
- Major Applications In Research And Healthcare
- Benefits, Challenges, And Key Considerations
Key Takeaways
| Point | Details |
|---|---|
| Digital PCR Precision | Digital PCR provides absolute quantification of nucleic acids, significantly enhancing sensitivity for detecting rare genetic variants compared to traditional methods. |
| Comparative Advantages | Unlike qPCR, which requires standard curves for quantification, digital PCR counts discrete positive and negative signals for accurate results. |
| Diverse Applications | Digital PCR is transformative in clinical diagnostics, oncology, and infectious disease monitoring, enabling detection and analysis previously deemed challenging. |
| Challenges to Consider | Despite its advantages, digital PCR poses operational complexities, including the need for specialized equipment and expertise, impacting cost and feasibility. |
Defining Digital PCR and Its Core Principles
Digital PCR (dPCR) represents a groundbreaking molecular biology technique that transforms traditional polymerase chain reaction (PCR) approaches by enabling unprecedented precision in nucleic acid quantification. Unlike conventional PCR methods that generate analog, exponential signals, digital PCR converts genetic amplification into discrete, countable digital signals, offering researchers an advanced method for absolute quantitation of target molecules.
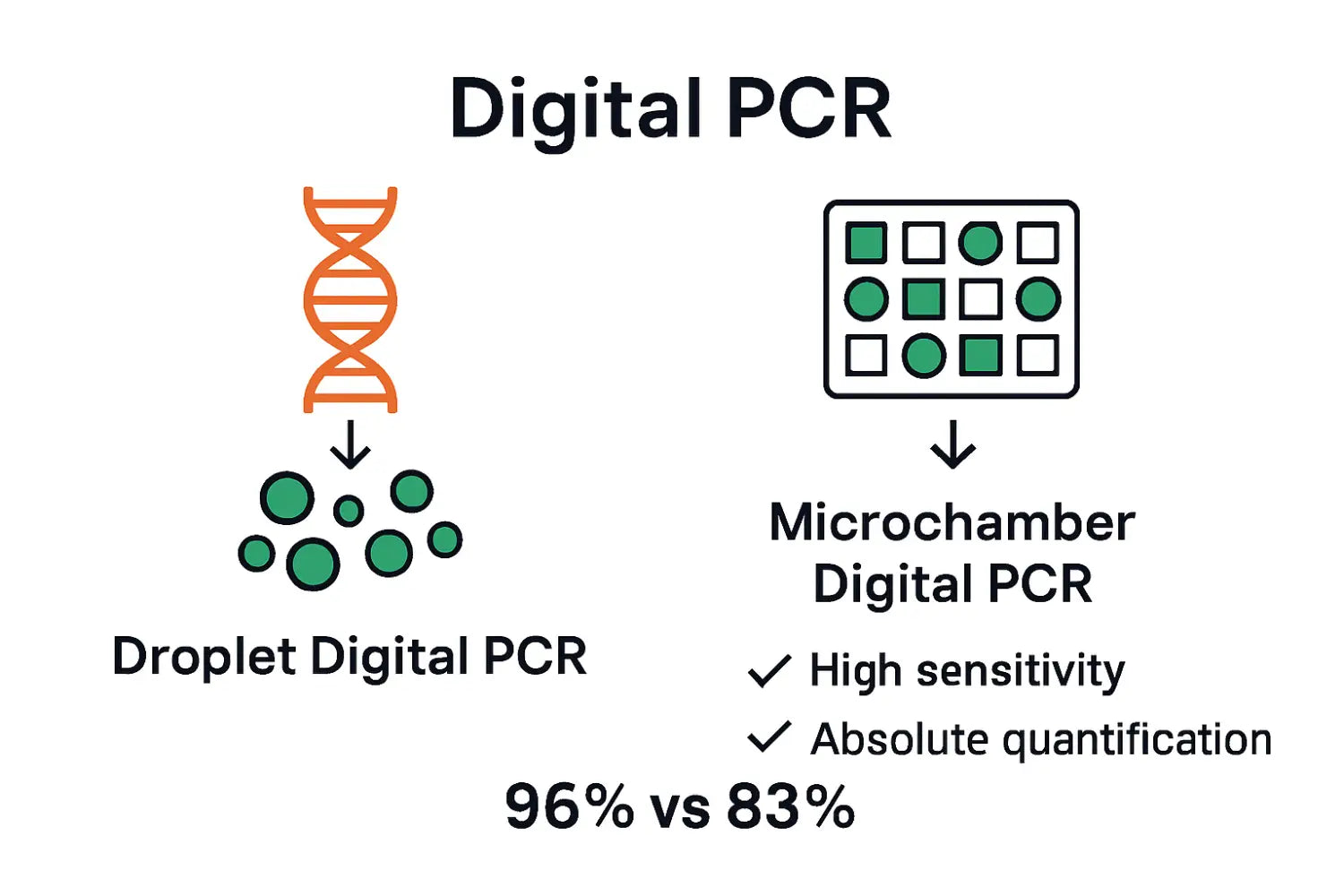
The fundamental principle of digital PCR involves strategically partitioning DNA samples into thousands of tiny reaction chambers or droplets, where each compartment contains either zero, one, or multiple template molecules. Droplet Digital PCR techniques allow scientists to statistically analyze genetic content with remarkable accuracy. By diluting samples and distributing them across multiple partitions, researchers can directly count positive and negative reactions, providing a direct measurement of target molecule concentration without relying on standard curves or reference standards.
Two primary digital PCR partitioning methodologies currently dominate scientific practice:
- Emulsion-based Digital PCR: Utilizes water-in-oil emulsion technology to create microscopic reaction chambers
- Microfluidics-based Digital PCR: Employs specialized chips with precisely engineered microchannels for sample distribution
These sophisticated approaches dramatically enhance molecular detection sensitivity, enabling researchers to identify and quantify rare genetic variants, detect low-frequency mutations, and perform highly precise genetic analysis across multiple scientific domains including clinical diagnostics, genetic research, and personalized medicine.
Comparing Digital PCR, qPCR, and Variants
Quantitative PCR (qPCR) and Digital PCR (dPCR) represent two powerful yet distinctly different molecular amplification techniques, each offering unique advantages for genetic analysis. While both methods enable nucleic acid detection and quantification, they fundamentally differ in their approach to signal measurement and statistical precision. Researchers select between these techniques based on specific experimental requirements, sample complexity, and desired quantification accuracy.

The primary distinction lies in their quantification strategies. qPCR generates continuous fluorescence signals that increase exponentially during amplification cycles, requiring standard curves or reference genes for relative quantification. In contrast, quantitative PCR technologies rely on interpolation and statistical inference. Digital PCR, however, transforms this paradigm by partitioning samples into thousands of individual reactions, allowing direct absolute quantification through binary positive/negative signal counting.
Key comparative characteristics between qPCR and dPCR include:
-
Quantification Method:
- qPCR: Relative quantification using fluorescence intensity
- dPCR: Absolute quantification via direct template counting
-
Sensitivity:
- qPCR: Limited sensitivity for rare variants
- dPCR: Superior sensitivity for detecting low-frequency mutations
-
Precision:
- qPCR: Higher variability, dependent on standard curves
- dPCR: Enhanced precision with reduced experimental variability
Beyond traditional qPCR and digital PCR, emerging variants continue to expand molecular diagnostics capabilities. Droplet digital PCR and microfluidic digital PCR represent advanced iterations that further refine partitioning techniques, offering researchers increasingly sophisticated tools for genetic analysis across clinical, research, and diagnostic applications.
Step-By-Step Digital PCR Workflow Explained
Digital PCR represents a sophisticated molecular biology technique that transforms genetic analysis through precise sample partitioning and amplification. The workflow demands meticulous preparation and execution, ensuring researchers can achieve unprecedented accuracy in nucleic acid quantification. Each stage requires careful attention to detail and specialized equipment to successfully implement this advanced technique.
The digital PCR workflow typically follows a structured sequence of critical steps. Laboratory workflow optimization becomes paramount when executing digital PCR, with researchers focusing on precise sample preparation and partition creation. The core stages include:
- Sample Preparation:
- Extract and purify target nucleic acids
- Assess sample quality and concentration
- Dilute samples to appropriate template density
- Partition Generation:
- Create microscopic reaction chambers
- Use either droplet-based or chip-based partitioning technologies
- Distribute DNA templates across thousands of individual reactions
- PCR Amplification:
- Add appropriate master mix and fluorescent probes
- Perform thermal cycling to amplify genetic material
- Ensure consistent reaction conditions across all partitions
- Signal Detection and Analysis:
- Count positive and negative partitions
- Analyze fluorescence signals
- Calculate absolute target molecule concentration
- Apply statistical methods to determine precise quantification
Advanced digital PCR workflows demand rigorous technical expertise, with researchers continuously refining techniques to maximize sensitivity and reproducibility across diverse scientific applications.
Major Applications in Research and Healthcare
Digital PCR has emerged as a transformative technology with profound implications across multiple scientific and medical domains. Its exceptional sensitivity and precision enable researchers and clinicians to detect and quantify genetic variations that were previously challenging or impossible to identify using traditional molecular techniques. The technology’s ability to provide absolute quantification of nucleic acid targets makes it invaluable in numerous critical applications.
In clinical diagnostics, digital PCR has revolutionized genetic screening and monitoring. Accessible research equipment has made these advanced techniques more available to laboratories worldwide, particularly in cancer research and precision medicine. Key clinical applications include:
-
Cancer Research:
- Detecting rare cancer mutations
- Monitoring treatment response
- Tracking minimal residual disease
- Analyzing circulating tumor DNA
-
Infectious Disease Diagnostics:
- Quantifying viral load
- Identifying low-frequency pathogens
- Monitoring antibiotic resistance genes
- Tracking emerging viral variants
Beyond clinical settings, digital PCR plays a crucial role in fundamental research and biotechnology. Researchers utilize this technique for precise gene expression analysis, rare mutation detection, and genomic research. In agricultural biotechnology, digital PCR helps scientists develop more resilient crop varieties by identifying and quantifying specific genetic traits with unprecedented accuracy.
The technology’s versatility extends to environmental monitoring, forensic science, and personalized medicine. By enabling detection of genetic variations at extremely low concentrations, digital PCR is pushing the boundaries of scientific understanding, offering researchers and clinicians powerful tools to investigate complex biological systems with remarkable precision.
Benefits, Challenges, and Key Considerations
Digital PCR represents a powerful molecular technique with significant advantages, yet it also presents unique challenges that researchers must carefully navigate. Understanding the nuanced landscape of digital PCR requires a comprehensive evaluation of its strengths, potential limitations, and critical implementation considerations.
The primary benefits of digital PCR are rooted in its unparalleled precision and sensitivity. High throughput screening approaches have been significantly enhanced by digital PCR’s ability to detect and quantify rare genetic variants with exceptional accuracy. Key advantages include:
-
Technical Benefits:
- Absolute quantification of nucleic acids
- Superior sensitivity for rare mutation detection
- Reduced impact of PCR amplification bias
- Direct measurement without standard curves
-
Operational Advantages:
- Increased reproducibility
- Lower sample volume requirements
- Enhanced statistical reliability
- Improved detection of low-frequency genetic variations
However, digital PCR is not without significant challenges. Technical complexities include sophisticated equipment requirements, higher per-sample costs, and the need for specialized training. Researchers must carefully optimize experimental design, manage partition generation techniques, and address potential sources of variability such as sample preparation quality and fluorescence detection parameters.
Implementation considerations extend beyond technical aspects, encompassing experimental design, cost-effectiveness, and institutional capabilities. Successful digital PCR applications demand a strategic approach that balances technological potential with practical constraints, ensuring researchers can leverage this powerful technique to advance scientific understanding across multiple disciplines.
Unlock Precision in Genetic Analysis with Accessible Digital PCR Solutions
Digital PCR offers unmatched sensitivity and absolute quantification for detecting rare mutations and low-frequency genetic variations. However, challenges such as sophisticated equipment requirements and experimental complexity can slow your progress in groundbreaking research or clinical diagnostics. If you are striving to enhance reproducibility while reducing costs and technical barriers, it is time to explore practical tools designed specifically to empower your workflow.
At Shop Genomics, we believe advanced molecular technologies like digital PCR should be within every researcher’s reach. We provide affordable, state-of-the-art research equipment that supports seamless sample partitioning, amplification, and accurate target quantification. Whether you are optimizing your laboratory workflow or pushing the boundaries of cancer mutation detection, our solutions help you achieve precise results without compromise.
Discover how you can accelerate your discoveries and overcome experimental challenges by visiting Shop Genomics. Learn more about optimizing your process in our guide on Laboratory Workflow Optimization and explore our Quantitative PCR Technologies for complementary molecular tools.

Take action today to transform your digital PCR research with accessible, reliable equipment designed for precision and efficiency. Unlock the future of genomic science at Shop Genomics.
Frequently Asked Questions
What is digital PCR?
Digital PCR (dPCR) is a molecular biology technique that provides precise quantification of nucleic acids by partitioning DNA samples into thousands of individual reaction chambers, allowing researchers to directly count positive and negative reactions for absolute quantification.
How does digital PCR differ from quantitative PCR (qPCR)?
The main difference is in their quantification methods: qPCR uses relative quantification based on fluorescence intensity and requires standard curves, while dPCR provides absolute quantification through direct counting of template molecules, offering higher sensitivity and precision.
What are the primary types of digital PCR?
The two primary types of digital PCR are emulsion-based digital PCR, which uses water-in-oil emulsion technology, and microfluidics-based digital PCR, which employs specialized chips with microchannels for sample distribution.
What applications benefit from using digital PCR?
Digital PCR is widely used in cancer research for detecting rare mutations, infectious disease diagnostics for quantifying viral load, and in agricultural biotechnology for precise genetic trait analysis. Its high sensitivity and accuracy make it essential in clinical diagnostics and fundamental research.




